The three-dimensional organization of the human genome (chromatin) is critically involved in the regulation of gene expression and is highly complex. It spans length scales from the radius of a DNA molecule (~1 nm) to 10 nm nucleosomes to chromatin domains (~100 nm) to chromosomal territories (~1µm) and to the cell nucleus (~10µm). Although the genetic and histone regulation of transcription can be addressed by molecular assays, decoding the role of chromatin packing requires nanoscale imaging. The spatial resolution of conventional microscopy is limited by the diffraction of light to approximately 200nm. In order to gain understanding of chromatin structure we are developing a new nanoscale imaging platform that can interrogate spatio-temporal changes in chromatin packing from the molecular level (e.g. individual DNA strands) to the whole chromatin in hundreds of live cells with sub-second temporal resolution in vitro and tissues in vivo, all providing nanoscale resolution of sensitivity.
The nanoimaging platform consists of a series of co-registered technologies: 3 nm resolution 3D chromatin scanning transmission electron microscopy (ChromSTEM), 6 nm resolution label-free superresolution microscopy (spectroscopic intrinsic contrast photolocalization optical nanoscopy, SICLON), 6 nm resolution multi-label spectroscopic stoch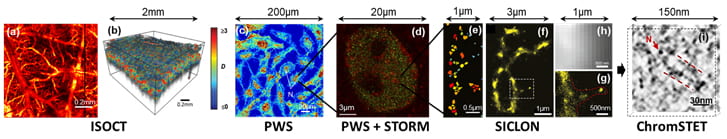 astic optical reconstruction microscopy (STORM), 20 nm sensitivity partial wave spectroscopic microscopy (PWS), and 40 nm sensitivity inverse spectroscopic optical coherence tomography (ISOCT). By combining and co-registering these methods we can optimize their synergy: label-free PWS provides the chromatin packing context, multi-label STORM shows the precise location of individual molecules without the danger of artifacts from heavy labeling, and electron microscopy tomography allows zooming in on the finest details of DNA organization.
astic optical reconstruction microscopy (STORM), 20 nm sensitivity partial wave spectroscopic microscopy (PWS), and 40 nm sensitivity inverse spectroscopic optical coherence tomography (ISOCT). By combining and co-registering these methods we can optimize their synergy: label-free PWS provides the chromatin packing context, multi-label STORM shows the precise location of individual molecules without the danger of artifacts from heavy labeling, and electron microscopy tomography allows zooming in on the finest details of DNA organization.
The imaging platform is complemented by state-of-the-art molecular and chromatin assays such as HiC and single cell sequencing to relate chromatin organization, its function, and gene expression regulation. In turn, the imaging data informs the predictive computational genomics modeling.
In addition to the chromatin nanoimaging platform, the Backman Lab has developed a platform of eight label-free nanoscale imaging technologies that together enable imaging from individual molecules to macroscopic tissue, including dual-band dual-scan ISOCT (D2ISOCT) for functional microangiography at the capillary scale, Low-coherence Enhanced Backscattering (LEBS) for assessment of tissue morphology based on the weak localization of light; Near-field Penetrating Optical Microscopy (NPOM); Structured Illumination OCT (SIOCT); Structured Illumination Laser Scanning Microscopy; Photonic Nanojet technology, which is now being widely used for a nanotechnology applications, Polarization-Gating Spectroscopy (PGS) for microvascular analysis, and finite-difference time-domain (FDTD) numerical solvers of Maxwell’s equations, which have enabled the first accurate computer modeling of essentially any type of microscopy without approximations.
Collaborators: Profs. Allen Taflove, Vinayak Dravid, Hao Zhang, Ji Yi
Partial Wave Spectroscopic Microscopy (PWS)
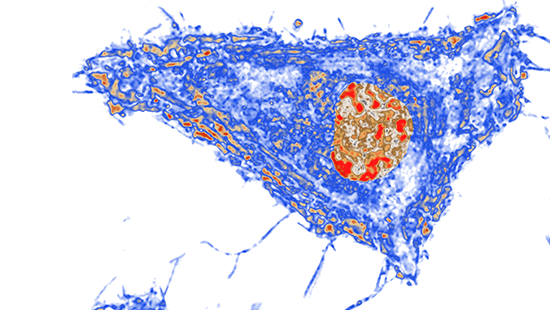
Partial-Wave Spectroscopic image of a cell. ‘Increase in chromatin packing scaling’ indicated in red.
Scaling properties of chromatin packing, which is a relationship between the genomic and physical sizes of chromatin polymer, plays a critical role in the chromatin regulation of gene expression. PWS enables imaging loci-specific scaling properties of chromatin packing in live cells, label-free, with sub-second temporal resolution, in hundreds of cells simultaneously, and with sensitivity to 20-350 nm length scales, which corresponds to genomic scales from the kbp to the Mbp range and topological scales from the fundamental chromatin chain to the topologically associated domains (TADs). The main principle of PWS is the detection of subdiffractional intracellular structures through the analysis of the interference of elastically scattered light scattered by these nanoscale structures. PWS has recently emerged as a powerful cell biology tool which was used to discover that alterations of chromatin packing scaling are one of the earliest events in carcinogenesis and its role in stem cell processes. The label-free PWS can be coupled with multi-label superresolution microscopy techniques such as STORM for localization of specific genes or other molecular species of interest involved in transcription with sub-10 nm resolution.
Collaborators: Prof. Allen Taflove
Highlight Papers: “Label-free imaging of the native, living cellular nano-architecture using partial-wave spectroscopic microscopy”, PNAS, 113(42) E6372-E6381 (2016). PMC5081614.
“Optical Methodology for Detecting Histologically Unapparent Nanoscale Consequences of Genetic Alterations in Biological Cells”, PNAS, 105(51), 20124–20129 (2008). PMC2629261.
Spectroscopic Intrinsic Contrast Photolocalization Optical Nanoscopy (SICLON)
The past two decades saw a major breakthrough in the emergence of super-resolution fluorescence microscopy with a Nobel Prize in Chemistry being awarded in 2014. However, the potential of this technology can still be enhanced. Existing methods rely on fluorescent labeling, which not only perturbs cell function, especially at the high label densities required for imaging dense structures such as chromatin, but also can onl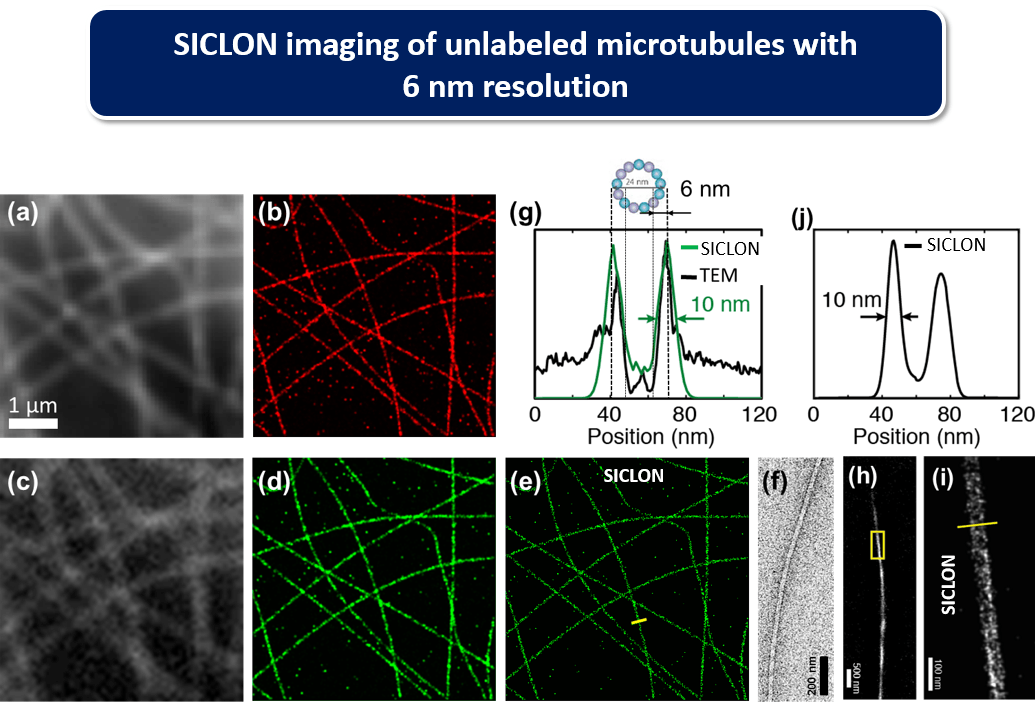 y be applied to known molecular species, and is difficult to implement quantitatively due to variable label uptake, diffusion and affinity.
y be applied to known molecular species, and is difficult to implement quantitatively due to variable label uptake, diffusion and affinity.
We have developed SICLON as a new technology for label-free and molecularly-specific imaging with sub-10 nm resolution. SICLON is based on a recently discovered physical effect: biopolymers such as DNA that until recently had been considered “dark” in the visible spectral range exhibit photoswitchable autoflorescence when illuminated by low-intensity visible light, which can be used for label-free nanoscopic imaging. SICLON combines time-resolved stochastic photon localization with simultaneous spectroscopic molecular-signature-carrying intrinsic fluorescence detection. SICLON couples the principles of single-molecule nanoscopy with two key innovations: endogenous fluorescence photoswitching effect, which bypasses fluorescence labeling, and a new technology able to simultaneously record not only the positions of emitting molecules but also their optical spectra, analysis of which enables SICLON to achieve molecular recognition and spatial resolution down to a few nanometers. SICLON is able to image unmodified DNA and chromatin without labeling with 6 nm spatial resolution.
Collaborators: Prof. Hao Zhang
Highlight Papers: “Sub-10nm imaging of nucleic acids using spectroscopic intrinsic-contrast photon-localization optical nanoscopy (SICLON)” Optics Letters, 43(23) 5817-5820 (2018).
“Parallel three-dimensional tracking of quantum rods using polarization-sensitive spectroscopic photon localization microscopy”, ACS Photonics, 4(7), 1747-1752 (2017).
Inverse Spectroscopic Optical Coherence Tomography (ISOCT)
It is becoming increasingly accepted that cell behavior in complex 3D tissue might be different from that of cells growing in culture. Chromatin imaging in tissue requires new technological solutions. ISOCT is an emerging technology that combines the 3D spatial resolution of OCT (~1-5 µm resolution and hundreds of microns depth of imaging) with spectral analysis of the signal from each resolution voxel to measure the statistics of subdiffractional macromolecular density arrangement inside the voxel with sensitivity down to 30 nm length scales. In particular, ISOCT is able to measure the scaling of chromatin packing density in cells located hundreds of microns below tissue surface as part of complex 3D tissue models (e.g. organoids, matrix models) or in situ / in vivo. The power of ISOCT lays in its capacity for fast (~1mm3/sec) in situ and in vivo imaging of tissue in 3D with micron resolution and nanoscale sensitivity, utilizing a low light power, and enabling fiber-optic probe implementation for in vivo use.
Collaborator: Prof. Ji Yi (Boston Medical Center)
Highlight Papers: “Spectral contrast optical coherence tomography angiography enables single-scan vessel imaging”Light: Science and Applications, 8(7) doi: 10.1038/s41377-018-0117-7 (2019).
“Single capillary oximetry and pericapillary ultrastructural sensing by dual-band dual-scan inverse spectroscopic optical coherence tomography” Light: Science and Applications, 7(57) DOI 10.1038/s41377-018-0057-2 (2018).
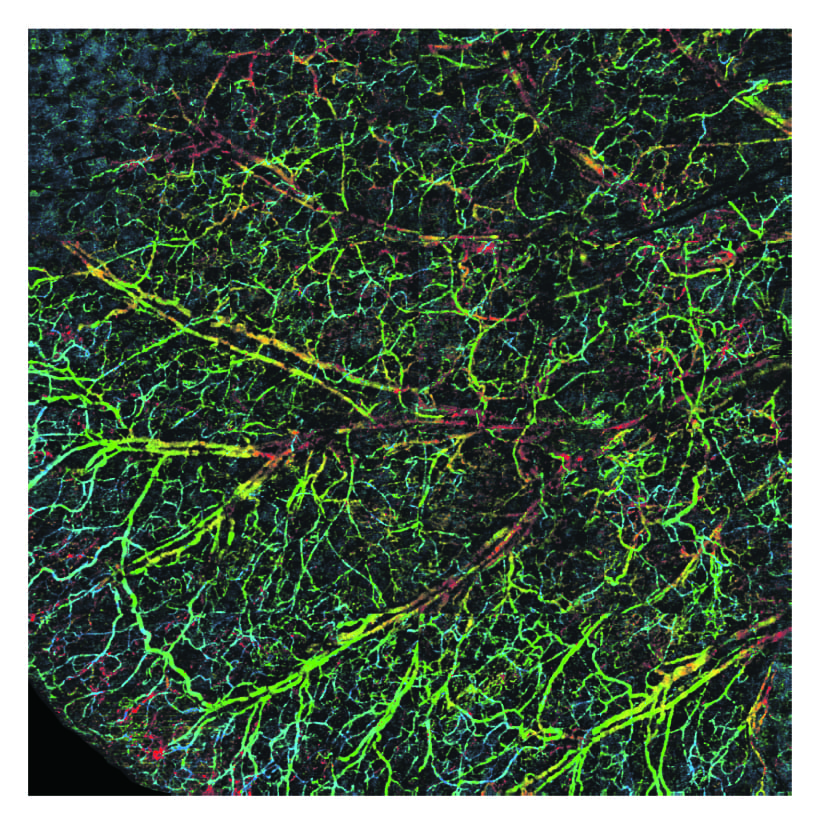
Spectral Contrast Optical Coherence Tomography Angiography (SC-OCTA)
Imaging capillaries is vital because it is the site where nutrient exchange happens between tissue and blood and thus can be indicative of disease and organ function. Unfortunately non-invasive imaging techniques, such as ultrasound, are limited in their resolution to larger blood vessels and as a result cannot tell how well blood is diffusing into capillary networks for nutrient exchange. We have developed a new technique called spectral contrast optical coherence tomography angiography (SC-OCTA) which can image capillaries with a simple scan of a non-harmful laser beam. This allows us to visualize capillary networks and pull out parameters such as tissue oxygen delivery. SC-OCTA works by performing 3D spectroscopy on tissue to locate capillaries, so unlike ultrasound or other optical speckle angiography techniques, its contrast is not affecting by low flow vessels or highly moving samples. This advantage is of crucial importance in endoscope imaging where you have a moving endoscope and moving sample. We are currently working to expand our technology into endoscope tips to enable deep endoscopy imaging for clinical applications.
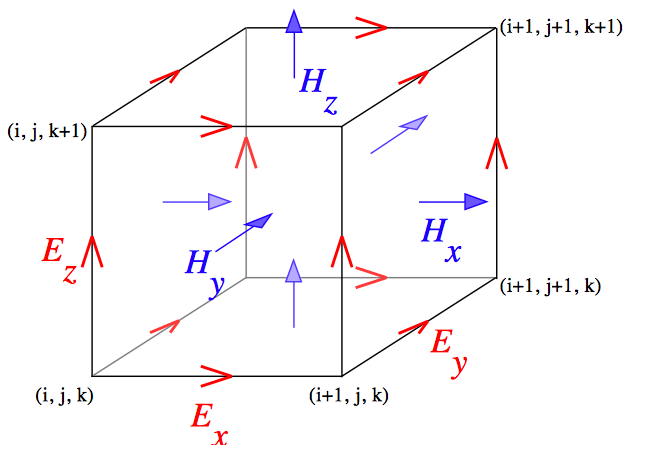
Computational Electrodynamics
Label-free optical imaging technologies such as PWS and ISOCT rely on elastic light scattering as their source of contrast. Light scattering is the most fundamental process underlying light-tissue interaction and is critical to a wide range of optical imaging and spectroscopy, from nano- and microscale cell microscopy to millimeter-scale tissue spectroscopy and to deep tissue imaging technologies such as diffuse optical tomography. Light scattering is generated by the spatial variations of intra- and extracellular density in tissue. Biological tissue is comprised of structures ranging from a few nanometers to tens of microns and all of these length scales may affect elastic scattering. Due to this extreme complexity of biological tissue, light scattering is poorly understood.
We develop robust computational and analytical methods in order to model light scattering in tissue. A key theme is that, from the electrodynamics perspective, biological tissue is modeled as a media with spatially continuous refractive index variations. Our computational electrodynamics platform enables first-principles modeling of essentially any type of optical microscopy based on any contrast mechanism including non-linear interactions, taking into account nanoscale details of realistic biological object, for objects from a few microns to millimeters in size, by accurately solving full-vector Maxwell’s equations without approximation. Key computational technologies are Finite-Difference Time-Domain (FDTD) and Pseudo-Spectral Time-Domain (PSTD) methods. We have developed an FDTD-based “microscope in a computer” software Angora, which is now available as open-source software. Angora solves Maxwell’s time-dependent equations with nanometer resolution within a specimen. FDTD computations are enabled by access to some of the most advanced supercomputers in the world.
Collaborators: Prof. Allen Taflove
Highlight Papers: “Interferometric spectroscopy of scattered light can quantify the statistics of subdifractional refractive-index fluctuations”, PRL, 111, 033903 (2013). PMC4123763
“The microscope in a computer: Image synthesis from three-dimensional full-vector solutions of Maxwell’s equations at the nanometer scale”, Progress in Optics, 57, 1-91 (2013)